Prediction and validation of circulating G-quadruplexes as a novel biomarker in colorectal cancer
Highlight box
Key findings
• The G-quadruplex (G4) is a novel biomarker in patients with colorectal cancer (CRC).
• The relationship between G4 and TP53 regulation may be a potential target.
What is known and what is new?
• It is well known that G4 plays an important role in the occurrence and development of tumors.
• The G4 is a novel biomarker in CRC because it is found for the first time in serum that CRC can be well distinguished from normal people.
What is the implication, and what should change now?
• We need to do more clinical samples and analyze the changes of G4 on more sequencing data.
Introduction
G-quadruplex (G4) is a noncanonical secondary structure consisting of four guanine-rich nucleic acid chains that can be formed from both DNA and RNA (1). Recent evidence suggests that G4 is involved in key genome functions, such as transcription, replication, genome stability, and epigenetic regulation, and has numerous connections to cancer biology, extending G4 beyond the DNA double helix (2). Although G4 structures can be computationally predicted from nucleic acid sequence motifs, their actual structures may vary significantly within and between motifs (3).
Analysis of G4’s secondary structures reveals that the endogenous G4 genome landscape is tightly regulated, with only 1–2% of the over 700,000 human sequences being able to biophysically fold into a G4 structure in vitro (4). It was reported that G4s have an increased prevalence in cancer cells (5) and are particularly associated with highly expressed and amplified genes in patient-derived aggressive breast cancer tissue (6). Multiple pieces of evidence suggest that G4 plays a role in cancer growth and progression (7,8), and higher G4 quantities have been detected in cancer cells compared to normal cells, making G4 an intriguing target for drug discovery (7,9). G4 appears to have an association with cancer-related genes, as a greater G4 is detected in cancer cells as compared to normal cells. The tracking of G4 in real time to directly understand its biological role is a new field of basic biology and may open up novel pathways for the diagnosis and treatment of diseases such as cancer (10). Moreover, the G4 structure may serve as a new prognostic biomarker and effective therapeutic target for colorectal cancer (CRC) (11).
We hypothesized that G4 content would significantly increase during the process of rapid cell proliferation, which is a hallmark of cancer. To test this hypothesis, we conducted bioinformatics analysis and identified changes in G4 content between tumor and normal cells. To confirm these findings, we performed experiments involving TP53 gene knockout and analyzed clinical samples. Our results revealed a marked increase in G4 content in tumor cells compared to normal cells, suggesting that G4 may serve as a promising diagnostic marker for cancer. Our findings shed light on the molecular mechanisms underlying tumorigenesis and suggest that G4 may play a significant role in cancer development. Further investigation into the role of G4 in tumor biology could lead to the development of novel therapeutic strategies for cancer treatment. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-26/rc).
Methods
Bioinformatics analysis of G4
We refer to a prior article for G4 predictions (4). There were five putative G4 sequences (PQSs) types to be predicted: (I) G3+L1_7; (II) G3+L1_12, (III) G2L1_12; (IV) G3+L8_12, and (V) G2+L1_12. In the related literature, there are several methods for predicting G4 sequences, including regular expression matching, scoring, sliding windows, and scoring and machine learning. We used the most classical type, regular expression matching (12).
The normal human genome was selected as the reference genome (ref; https://www.ncbi.nlm.nih.gov/genome/guide/human/) and the sequence of cell line SW480 of CRC (mut) was obtained from the DepMap database of the Broad Institute (https://depmap.org/portal/). The mutation [including single-nucleotide polymorphism (SNP) and insertion-deletion (INDEL)] position of the SW480 cells was obtained, and a sequence of 50 nt was slid up and down from the human reference genome (hg19; since the mutation information was based on hg19) to construct a normal reference sequence. The corresponding position was then replaced with a mutation site or sequence. Thus, the mutation sequence was constructed and processed, and the two formed a reference-mutation sequence pair (13). A program was written to obtain the PQS of the reference sequence and mutation sequences by regular matching, including the positive and negative chains. After the PQSs were obtained, statistical analysis was performed. The Fisher exact test was used to compare whether the same mutation type showed obvious differences between reference and mutation sequences, and the PQS count number was compared here.
Cell lines and culture
One common human CRC cell line (SW480) was obtained from Suzhou Haixing Biosciences (Suzhou, China), and one normal human colon cell line (NCM460) was obtained from Peking University Health Science Center (Beijing, China). All cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with 12.5 mM glucose, 4 mM glutamine, 5% fetal bovine serum (10270; Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and 1% streptomycin/penicillin at 37 ℃ in a 5% CO2 atmosphere.
MTT assay
Cells seeded in 96-well plates at a density of 5×103 cells per well were incubated at 37 ℃ in a 5% CO2 atmosphere for 48 h. The medium was removed, and cells were then incubated with 100 µL of a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma-Aldrich, St. Louis, MO, USA) solution (5 mg/mL) for 2–4 h. After the medium was discarded, 100 µL of dimethyl sulfoxide (DMSO) was added and gently mixed for 10 min. Finally, the absorbance at 570 nm was measured by a microplate reader (Synergy H1, BioTek, Winooski, VT, USA). Three replicates per condition were assayed, and the average values from three to five separate experiments are presented. Data are expressed as a percentage of the control.
TP53 gene CRISPR/Cas9 knockout
We used CRISPR/Cas9 to knock out the TP53 gene (gene ID: 7157) in SW480 cells. A pX330 p53 vector (plasmid #42230; Addgene, Watertown, MA, USA) was used to knock out human mutant TP53 (14). SW480 cells were transfected with pX330 vector harboring single-guide RNA (sgRNA) specific to human TP53 using FugeneHD reagent (Promega Corp., Madison, WI, USA) (sgRNA sequence: 5'-CTTCCCACAGGTCTCTGCTA-3'). After 48 h, single cells were seeded on 96 well plates, and upon clone expansion, TP53 expression levels were examined by polymerase chain reaction (PCR), gel electrophoresis, and sequencing.
Serum collection and patient information
Serum samples (n=27) were obtained from patients who underwent surgical resection of primary CRC between 2020 and 2022 at Peking University People’s Hospital. Normal samples were obtained from healthy volunteers undergoing routine physical examinations at the medical examination center. We collected 5 mL of blood using ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes and collected 1 mL of serum for the experiment. The research protocol was approved by the Ethics Committee of Peking University People’s Hospital (approval No. 2018PHB193-01), and all patients provided appropriate informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The CRC samples were obtained from patients who had been clinically diagnosed and operated, while the control samples were collected from the serum of healthy individuals at the physical examination center. Detailed clinical data can be found in Table S1. The study included 27 CRC patients (Tumor) and 27 healthy individuals (Normal) (refer to Table S1). There were no significant differences observed in age and sex between the patients and healthy individuals.
Measurement of G4
The DNA samples were extracted using TransGen Biotech kits (Beijing, China), following the manufacturer’s instructions. The formation of G4 was assessed by measuring Thioflavin T (ThT) fluorescence spectroscopy (15-19). We used the sequence (5'-GGGTTAGGGTTAGGGTTAGGG-3') of the G4 structure of human telomeres as the G4 standard sample (G4 standard) using gene synthesis. Before analysis, the G4 standard was heated at 90 ℃ for 5 min at a 10-µM concentration in water, then diluted into 100 mM Tris-HCl (pH 7.5, 10 mM KCl, 5 mM MgCl2), and slowly cooled to room temperature (22–25 ℃) over 1 h. Experiments were performed using 96-well black microplates from BBI Life Sciences Corporation (Shanghai, China). Each condition was tested at least in triplicate in a volume of 20 µL for each DNA sample. Measurements were taken at room temperature (22–25 ℃). The G4 standard and ThT were mixed at final concentrations of 1 and 0.5 mM, respectively. Fluorescence emission was detected at 490 nm after excitation at 440 nm in a microplate reader (Synergy H1, BioTek). Experiments were performed under the same conditions as used for the fluorescence single-wavelength measurements except that the fluorescence emission was collected between 460 and 600nm every 5 nm with a microplate reader (Synergy H1, BioTek).
Statistical analysis
A t-test was performed to calculate the P value. A histogram was plotted using GraphPad Prism v. 8.8 (GraphPad Software, Inc., La Jolla, CA, USA), and the receiver operating characteristic (ROC) curve was plotted using R language (The R Foundation of Statistical Computing). Data are expressed as the mean ± standard deviation.
Results
Bioinformatics analysis of G4
Quantitative prediction of PQS
There were 1,231,281 mutation sites in SW480 cells, including 1,147,609 SNPs, 30,052 insertions (INSs), 52,996 deletions (DELs), 622 double-nucleotide polymorphisms, and 2 triple-nucleotide polymorphisms. Reference-mutation sequence pairs were generated for these mutation sites, and the PQS was predicted from these sequences (Figure 1).
Quantitative prediction of PQS and non-PQS
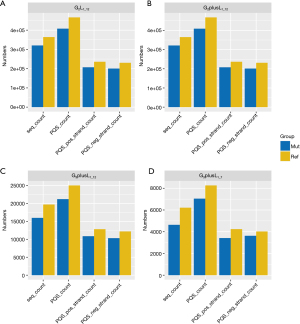
We examined the number of original PQS mutations that underwent conversion to non-PQS mutations (defined as S1) and the number of original non-PQS mutations that were converted to PQS mutations (defined as S2) following sequence mutation. From Figure 2, the S1 number is higher than that of S2, indicating that mutation did not increase the PQS number.
Differences between the sequence pairs of the different mutation types
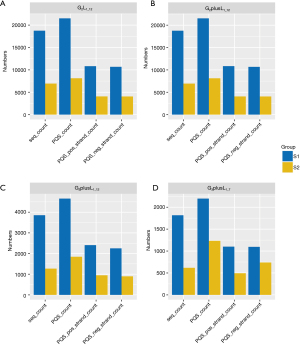
We categorized the mutations into different types and analyzed the differences between PQS in the reference-mutation sequence pairs. Annotation information for the mutation was obtained from the DepMap database. The database annotates mutations (mut_anno) into four categories: damaging, silent, other preserving, and nonconserving. Additionally, mutations were classified into 20 classes (Mut_Class) according to the database terms, including 3'-UTR, 5'-FLANK, 5'-UTR, DE_NOVO_start_out of FRAME, FRAME_SHIFT_DEL, FRAME_SHIFT_INS, IGR, IN_FRAME_DEL, In_Frame_Ins, Intron, Missense_Mutation, Nonsense_Mutation, Nonstop_Mutation, Silent, Splice_Site, Start_Codon_Del, Start_Codon_Ins, Start_Codon_SNP, Stop_Codon_Del, and Stop_Codon_Ins. Based on the above classifications, the statistical analysis results of the mutation sharing into Mut_Anno are shown in Figure 3. The reference group was still mostly higher because the reference group itself predicts a higher PQS.
Fisher exact test of PQS
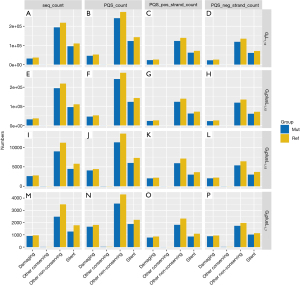
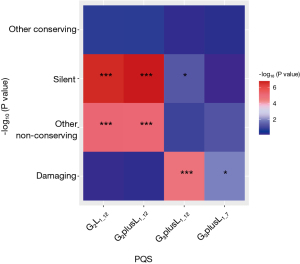
Fisher exact test requires the construction of contingency tables. With the damaging of G2L1_12 used as an example, the constructed contingency tables are shown in Table 1. As shown in Figure 4, the count numbers of reference and mutation vary significantly among different PQSs. This difference is reflected in the silent, other nonconserving sequences of G2L1_12 and G2plusL1_12. The damaging sequence showed the most significant difference in G3plusL1_12.
Table 1
| G2L1_12 type | Reference | Mutation |
|---|---|---|
| Damaging | 52,481 | 46,008 |
| Nondamaging | 274,487+142,935+244 | 242,046+122,937+242 |
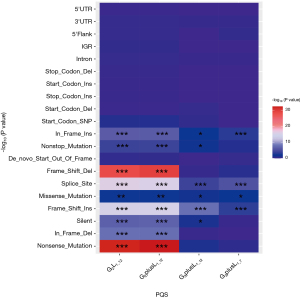
Mut_class was analyzed in the same way as above, and the result of the Fisher exact test is shown in Figure 5. This result indicated that there are significant differences between G2L1_12 and G2plusL1_12, including In_Frame_In, Nonstop_Mutation, De-novo-Start_OutFrame, Frame_Shift_Del, Splice_Site, Missense Mutation, Frame_Shift_Ins, Silent, In-Frame_Del, and Nonsense_Mutation. However, in G3plusL1_12 and G3plusL1_7, Splice_Site and Frame_Shift_Ins were significantly different. This indicated that the different types of G4 varied in terms of mutations and were mainly focused on the G2L1_12 and G2plusL1_12 types, because the types have more differences.
PQS analysis of tumor genes
Tumor gene data were obtained from ONGene (www.ongene.bioinfo-minzhao.org). There were 803 tumor genes, 690 of which were coincident with the genes in the SW480 mutation data. The difference between tumor and nontumor genes and between reference-mutation sequence pairs was analyzed first. The results are shown in Table 2.
Table 2
| G4 classes | Sequence number | PQS number | − | + |
|---|---|---|---|---|
| G2L1_12 | 0.3738 | 0.0671 | 0.2343 | 0.1559 |
| G2+L1_12 | 0.3440 | 0.0530 | 0.2556 | 0.1048 |
| G3+L1_12 | 0.5161 | 0.0567 | 0.0544 | 0.4055 |
| G3+L1_7 | 0.7332 | 0.0109 | 0.0489 | 0.0635 |
The P values for the Fisher exact test, which indicates that the classical PQS G3+L1_7 had significant differences between neoplastic and nonneoplastic genes (P=0.0109). PQS, putative G-quadruplex sequence; −, negative strand; +, positive strand.
Knockout of TP53 gene by CRISPR/Cas9
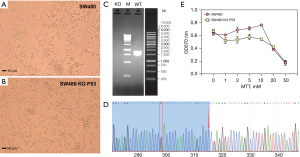
We used CRISPR/Cas9 to knock out the TP53 gene in SW480 cells. The results showed that after knockout of the TP53 gene, the growth state of the cells was changed and that the cell appearance was small (Figure 6A,6B). Moreover, agarose gel electrophoresis after PCR amplification clearly indicated that TP53 was not present in the knockout samples (KO) (Figure 6C). The deletion of key bases was found after Sanger sequencing of the PCR product (Figure 6D). The MTT experiment also showed that KO had low cell activity and were significantly different from nonknockout cells (SW480 cell) (Figure 6E).
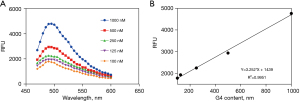
The fluorescence spectrum and the standard curve of G4
We used G4 standard with ThT reaction after each 5 nm fluorescence spectrum scan (excitation wavelength 440 nm, emission wavelength range 460–600 nm) and found the maximum absorption of fluorescence value at 490 nm (Figure 7A). Figure 7B shows the standard and the sample for the determination of 490 nm, the obtained fluorescence value, the concentration of the standard by linear regression to produce a standard curve, and the concentration of the sample calculated according to the equation of the standard curve.
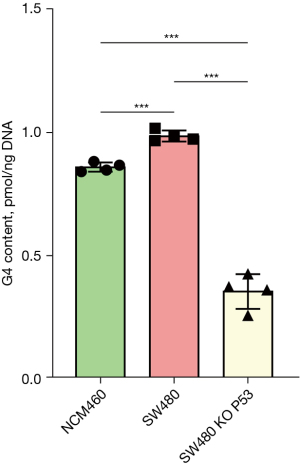
G4 contents were significantly different between NCM460, SW480, and KO TP53 SW480 cells
G4 content was detected via the ThT fluorescence method. The results showed that the G4 content in SW480 cells was significantly higher than that in the NCM460 cells. When the TP53 gene was knocked out in SW480 cells, the G4 content decreased significantly, from 0.99 to 0.36 (pmol/ng DNA), representing a decrease of 64% (Figure 8).
G4 as a candidate biomarker in CRC diagnosis
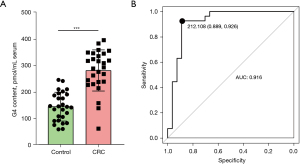
We measured the serum G4 content in 27 healthy participants and 27 patients with CRC. The results showed that the G4 content in patients with CRC was significantly higher (a 1.94-fold increase) than that in healthy participants (P<0.001), with an average increased from 144.6229 to 281.0693 pmol/mL serum (Figure 9A). ROC evaluation using this result showed that the area under the ROC curve (AUC) was 0.916, with good specificity and sensitivity of 0.889 and 0.926, respectively (Figure 9B). Therefore, G4 may be considered a potential biomarker of CRC.
Discussion
G4 is a unique genomic structure composed of multiple guanine nucleotides that plays a significant role in various biological processes. In recent years, emerging evidence has linked G4 formation to tumorigenesis, making it a promising target for cancer treatment (20-24). Our study aimed to explore the relationship between G4 formation and tumor development and to investigate the therapeutic potential of targeting G4 structures in cancer treatment (7,25,26). Through this research, we hope to shed light on the molecular mechanisms underlying cancer progression and contribute to the development of novel therapeutic strategies.
Although G4 prediction studies have demonstrated the relevance of G4 structures, the specific quantity of G4 structures present in tumor cells remains uncertain (12,27-31). Our study sought to address this deficiency by characterizing the abundance of G4 structures in cancer cells and assessing their potential as a therapeutic target. In this study, based on a previous prediction method, we accurately predicted the G4 quantity of SW480 cells in CRC and compared it with the reference genome. We found that the G4 quantity of SW480 did not increase significantly but decreased slightly. Moreover, the changes of G4 content in CRC cells have not been reported. We thus detected the G4 content of DNA in cells and human serum using ThT fluorescence labeling and found that the G4 content in CRC tumor cells was higher than that in normal human cells. To determine the relationship between G4 and tumor, we knocked out the TP53 gene which was closely related to the tumor. We found that after the TP53 gene was knocked out, the G4 content was decreased significantly and was much lower than that in SW480 without TP53 gene knockout and in normal cells. This suggests that G4 is closely related to tumor occurrence and that TP53 could increase the G4 content in cells. This result is consistent with studies of other cancers, such as breast cancer (6), liver cancer, stomach cancer (5), and lung cancer (32). In a study, G4 has been used as a novel target for the treatment of gastrointestinal cancers and in the treatment of esophageal cancer, pancreatic cancer, hepatocellular carcinoma, gastric cancer, CRC, and gastrointestinal stromal tumors (33). However, the regulation of G4 and ligand function in cancer is not clear. Therefore, it is necessary to explore new modulations, such as the direct or indirect interactions with TP53 and G4 (34-36). TP53 might be an important regulator or target of the G4 (34) may lead to a significant reduction of transcriptional activity and change in the dynamics of p53α isoforms (37). It may also increase the transactivation ability of partial function TP53 family proteins (38). Nevertheless, the other findings indicate that p53 expression did not have a prognostic impact on sporadic CRCs in Korean patients (39). Furthermore, it has been discovered that berberine may inhibit colon cancer by regulating the TCA cycle and glycolysis/gluconeogenesis through its interaction with c-MYC and HIF1α G-quadruplexes (40). Here, our results showed that TP53 knockout significantly decreased G4 content. We can judge the prognosis effect by detecting the change of serum G4 content. With the continuous decline of G4 content, the prognosis effect may be better and better. Therefore, G4 can be used not only for diagnosis, but also for judging the prognosis of CRC.
We predicted G4 quantity in the genome of SW480 cell lines, detected G4 content in cell and human serum, and found increased G4 levels in CRC. G4 content and TP53 expression were positively correlated in SW480 cells. The G4 and TP53 regulatory relationship might be a potential target for cancer research in the future. With increased research attention in this area, the related mechanisms will be better understood, hopefully to the benefit of those with cancer.
Conclusions
For the first time, we predicted the number and distribution of G4 in the SW480 human CRC cell line, with the predicted result being significantly different from the experimental result. We surmised that this difference may be attributable to environmental differences in the cells and the structures of the genome. Finally, we found that the G4 content in patients with CRC was significantly increased compared to healthy controls. Via the model, we determined that G4 is a good biomarker for the diagnosis of CRC, and, for the first time, confirmed that G4 has diagnostic potential in CRC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-26/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-26/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-26/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-26/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Varshney D, Spiegel J, Zyner K, et al. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol 2020;21:459-74. [Crossref] [PubMed]
- Spiegel J, Adhikari S, Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem 2020;2:123-36. [Crossref] [PubMed]
- Maizels N. G4-associated human diseases. EMBO Rep 2015;16:910-22. [Crossref] [PubMed]
- Marsico G, Chambers VS, Sahakyan AB, et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res 2019;47:3862-74. [Crossref] [PubMed]
- Biffi G, Tannahill D, Miller J, et al. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One 2014;9:e102711. [Crossref] [PubMed]
- Hänsel-Hertsch R, Simeone A, Shea A, et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet 2020;52:878-83. [Crossref] [PubMed]
- Kosiol N, Juranek S, Brossart P, et al. G-quadruplexes: a promising target for cancer therapy. Mol Cancer 2021;20:40. [Crossref] [PubMed]
- Asamitsu S, Obata S, Yu Z, et al. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019;24:429. [Crossref] [PubMed]
- Monsen RC, Maguire JM, DeLeeuw LW, et al. Drug discovery of small molecules targeting the higher-order hTERT promoter G-quadruplex. PLoS One 2022;17:e0270165. [Crossref] [PubMed]
- Di Antonio M, Ponjavic A, Radzevičius A, et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat Chem 2020;12:832-7. [Crossref] [PubMed]
- Wu R, Li L, Bai Y, et al. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death Dis 2020;11:908. [Crossref] [PubMed]
- Puig Lombardi E, Londoño-Vallejo A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res 2020;48:1-15. [Crossref] [PubMed]
- Gong JY, Wen CJ, Tang ML, et al. G-quadruplex structural variations in human genome associated with single-nucleotide variations and their impact on gene activity. Proc Natl Acad Sci U S A 2021;118:e2013230118. [Crossref] [PubMed]
- Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014;514:380-4. [Crossref] [PubMed]
- Lv L, Zhang L. Characterization of G-Quadruplexes in Enterovirus A71 Genome and Their Interaction with G-Quadruplex Ligands. Microbiol Spectr 2022;10:e0046022. [Crossref] [PubMed]
- Renaud de la Faverie A, Guédin A, Bedrat A, et al. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res 2014;42:e65. [Crossref] [PubMed]
- Zhang S, Sun H, Chen H, et al. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim Biophys Acta Gen Subj 2018;1862:1101-6. [Crossref] [PubMed]
- Xu S, Li Q, Xiang J, et al. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci Rep 2016;6:24793. [Crossref] [PubMed]
- Liu S, Peng P, Wang H, et al. Thioflavin T binds dimeric parallel-stranded GA-containing non-G-quadruplex DNAs: a general approach to lighting up double-stranded scaffolds. Nucleic Acids Res 2017;45:12080-9. [Crossref] [PubMed]
- Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J 2010;277:1107-17. [Crossref] [PubMed]
- Festa C, Esposito V, Benigno D, et al. Discovering New G-Quadruplex DNA Catalysts in Enantioselective Sulfoxidation Reaction. Int J Mol Sci 2022;23:1092. [Crossref] [PubMed]
- Wang SK, Su HF, Gu YC, et al. Complicated behavior of G-quadruplexes and evaluating G-quadruplexes' ligands in various systems mimicking cellular circumstance. Biochem Biophys Rep. 2015;5:439-47. [Crossref] [PubMed]
- Bhatt U, Kretzmann AL, Guédin A, et al. The role of G-Quadruplex DNA in Paraspeckle formation in cancer. Biochimie 2021;190:124-31. [Crossref] [PubMed]
- Kim S, Hwang S. G-Quadruplex Matters in Tissue-Specific Tumorigenesis by BRCA1 Deficiency. Genes (Basel) 2022;13:391. [Crossref] [PubMed]
- Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol 2017;18:279-84. [Crossref] [PubMed]
- Nakanishi C, Seimiya H. G-quadruplex in cancer biology and drug discovery. Biochem Biophys Res Commun 2020;531:45-50. [Crossref] [PubMed]
- Brázda V, Kolomazník J, Lýsek J, et al. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics 2019;35:3493-5. [Crossref] [PubMed]
- Mishra SK, Tawani A, Mishra A, et al. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep 2016;6:38144. [Crossref] [PubMed]
- Wu F, Niu K, Cui Y, et al. Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol 2021;4:98. [Crossref] [PubMed]
- Yadav VK, Abraham JK, Mani P, et al. QuadBase: genome-wide database of G4 DNA--occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res 2008;36:D381-5. [Crossref] [PubMed]
- Dhapola P, Chowdhury S. QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res 2016;44:W277-83. [Crossref] [PubMed]
- Liu LY, Liu W, Wang KN, et al. Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination. Angew Chem Int Ed Engl 2020;59:9719-26. [Crossref] [PubMed]
- Han ZQ, Wen LN. Application of G-quadruplex targets in gastrointestinal cancers: Advancements, challenges and prospects. World J Gastrointest Oncol 2023;15:1149-73. [Crossref] [PubMed]
- Zhang L, Lu Y, Ma X, et al. The potential interplay between G-quadruplex and p53: their roles in regulation of ferroptosis in cancer. Front Mol Biosci 2022;9:965924. [Crossref] [PubMed]
- Solomon H, Dinowitz N, Pateras IS, et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 2018;37:1669-84. [Crossref] [PubMed]
- Marcel V, Tran PL, Sagne C, et al. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 2011;32:271-8. [Crossref] [PubMed]
- Porubiaková O, Bohálová N, Inga A, et al. The Influence of Quadruplex Structure in Proximity to P53 Target Sequences on the Transactivation Potential of P53 Alpha Isoforms. Int J Mol Sci 2019;21:127. [Crossref] [PubMed]
- Vojsovič M, Kratochvilová L, Valková N, et al. Transactivation by partial function P53 family mutants is increased by the presence of G-quadruplexes at a promoter site. Biochimie 2024;14-23. [Crossref] [PubMed]
- Kim HR, Kim HC, Yun HR, et al. An alternative pathway in colorectal carcinogenesis based on the mismatch repair system and p53 expression in Korean patients with sporadic colorectal cancer. Ann Surg Oncol 2013;20:4031-40. [Crossref] [PubMed]
- Wen L, Han Z, Li J, et al. c-MYC and HIF1α promoter G-quadruplexes dependent metabolic regulation mechanism of berberine in colon cancer. J Gastrointest Oncol 2022;13:1152-68. [Crossref] [PubMed]